SPARQL
- A query language adapted for the structure of the data necessary!
- Enter SPARQL
- SPARQL Protocol And RDF Query Language
- Similar syntax as SQL
- But uses pattern matching on graphs instead of joins over tables
Leif Harald Karlsen
species.description:
sid | genus | common_name | global_conservation | local_conservation | blacklisted
-----+-----------------------+----------------+---------------------+--------------------+-------------
1 | Rattus Norvegicus | Brown rat | 6 | 6 | f
2 | Larus Canus | Common gull | 6 | 6 | f
3 | Lutra | Eurasian otter | 5 | 4 | f
4 | Osmoderna eremita | Hermit beetle | 5 | 4 | f
5 | Malacodea regelaria | | | 3 | f
6 | Balaenoptera physalus | Fin whale | 4 | 6 | f
7 | Emberiza calandra | Corn bunting | 6 | 0 | f
8 | Stelis phaeoptera | | | 0 | f
9 | Arion vulgaris | Spanish slug | 6 | 6 | t
10 | Lupinus polyphyllus | garden lupin | 6 | 6 | tspecies.observation:
oid | species | observed_time | observed_lat | observed_long | location
-----+---------+---------------------+--------------+---------------+------------
1 | 1 | 2020-10-21 02:14:10 | 59.89 | 10.72 |
2 | 3 | 2021-01-01 02:14:10 | 59.39 | 10.78 |
3 | 5 | 2020-11-03 02:14:10 | 59.47 | 11.02 |
4 | 2 | 2020-01-17 02:14:10 | 58.91 | 10.34 |
5 | 6 | 2019-12-24 02:14:10 | 59.88 | 11.01 |
6 | 1 | 2020-06-09 02:14:10 | | | Oslo
7 | 1 | 2021-01-02 02:14:10 | 59.81 | 9.94 |
8 | 10 | 2020-04-30 02:14:10 | 59.11 | 10.7 |
9 | 2 | 2019-02-12 02:14:10 | 59.36 | 11.08 |
10 | 1 | 2020-08-22 02:14:10 | 59.89 | 9.28 |
11 | 10 | 2020-10-20 02:14:10 | 60.01 | 10.78 |
12 | 9 | 2020-03-26 02:14:10 | 59.37 | 10.23 |
13 | 2 | 2020-09-04 02:14:10 | 59.41 | 10.4 |
14 | 1 | 2019-12-03 02:14:10 | 59.39 | 11.04 |
15 | 10 | 2020-10-06 02:14:10 | 59.11 | 10.83 |
16 | 2 | 2020-01-03 02:14:10 | 59.19 | 9.31 |
17 | 9 | 2020-07-07 02:14:10 | 59.64 | 10.61 |
18 | 10 | 2020-04-19 02:14:10 | 59.23 | 10.39 |
19 | 1 | 2019-03-08 02:14:10 | | | Drammen
20 | 2 | 2021-01-21 02:14:10 | | | Oslo
21 | 8 | 2020-11-30 02:14:10 | 58.99 | 9.78 |
22 | 9 | 2020-10-09 02:14:10 | 59.83 | 10.37 |
23 | 1 | 2020-07-23 02:14:10 | | | Nord-markaspecies.eats:
sid1 | sid2
------+------
1 | 4
3 | 4
2 | 1
9 | 10
2 | 9
9 | 4
4 | 10WITH -- named subqueries
SELECT -- project and manipulate values
FROM -- choose tables the query should use (and join them together)
WHERE -- filter rows
GROUP BY -- group rows together (for group-wise aggregation)
HAVING -- filter groups
ORDER BY -- order rows in result
OFFSET -- skip rows
LIMIT -- limit to first number of rowsSee the PostgreSQL documentation for more info on any of these.
Find the common name of all blacklisted species:
Find the common and scientific names and time of all observations of
all species having conservation status less than 6 either
locally or globally:
Find out how many times each species have been observed.
Only those that are observed at least once:
SELECT d.genus
FROM species.description AS d1
JOIN species.eats AS e ON (e.s1 = d1.sid)
JOIN species_description AS d2 ON (e.s2 = d2.sid)
WHERE d2.common_name = 'Hermit beetle'
UNION
SELECT d.genus
FROM species.description AS d1
JOIN species.eats AS e1 ON (e1.s1 = d1.sid)
JOIN species.eats AS e2 ON (e1.s2 = e2.s1)
JOIN species_description AS d2 ON (e2.s2 = d2.sid)
WHERE d2.common_name = 'Hermit beetle'
UNION
SELECT d.genus
FROM species.description AS d1
JOIN species.eats AS e1 ON (e1.s1 = d1.sid)
JOIN species.eats AS e2 ON (e1.s2 = e2.s1)
JOIN species.eats AS e3 ON (e2.s2 = e3.s1)
JOIN species_description AS d2 ON (e3.s2 = d2.sid)
WHERE d2.common_name = 'Hermit beetle'
...eats!Simple example: numbers
Transitive closure of eats:
WITH RECURSIVE -- note RECURSIVE-keyword!
tc(sid1, sid2) AS (
SELECT sid1, sid2 -- base case
FROM species.eats
UNION ALL
SELECT e.sid1, tc.sid2
FROM species.eats AS e
JOIN tc ON (e.sid2 = tc.sid1) -- recursive query
)
SELECT *
FROM tc;General transitive closure of T:
Who transitively eats Hermit beetles?
WITH RECURSIVE
tc(sid1, sid2) AS (
SELECT sid1, sid2
FROM species.eats
UNION ALL
SELECT e.sid1, tc.sid2
FROM species.eats AS e
JOIN tc ON (e.sid2 = tc.sid1)
)
SELECT DISTINCT d1.genus
FROM species.description AS d1
JOIN tc ON (tc.sid1 = d1.sid)
JOIN species.description AS d2 ON (tc.sid2 = d2.sid)
WHERE d2.common_name = 'Hermit beetle';RECURSIVE-keyword is mandatory in PostgreSQL, but
optional in some other RDBMSs@prefix exd: <http://example.org/species/desc/> .
@prefix exo: <http://example.org/species/obs/> .
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .
exd:1 a exd:Species;
exd:genus "Rattus Norvegicus";
exd:common_name "Brown rat";
exd:global_conservation 6;
exd:local_conservation 6;
exd:blacklisted false ;
exd:eats exd:4 .
exd:2 a exd:Species;
exd:genus "Larus Canus";
exd:common_name "Common gull";
exd:global_conservation 6;
exd:local_conservation 6;
exd:blacklisted false ;
exd:eats exd:1, exd:9 .
exd:3 a exd:Species;
exd:genus "Lutra";
exd:common_name "Eurasian otter";
exd:global_conservation 5;
exd:local_conservation 4;
exd:blacklisted false ;
exd:eats exd:4 .
exd:4 a exd:Species;
exd:genus "Osmoderna eremita";
exd:common_name "Hermit beetle";
exd:global_conservation 5;
exd:local_conservation 4;
exd:blacklisted false ;
exd:eats exd:10 .
exd:5 a exd:Species;
exd:genus "Malacodea regelaria";
exd:local_conservation 3;
exd:blacklisted false .
exd:6 a exd:Species;
exd:genus "Balaenoptera physalus";
exd:common_name "Fin whale";
exd:global_conservation 4;
exd:local_conservation 6;
exd:blacklisted false .
exd:7 a exd:Species;
exd:genus "Emberiza calandra";
exd:common_name "Corn bunting";
exd:global_conservation 6;
exd:local_conservation 0;
exd:blacklisted false .
exd:8 a exd:Species;
exd:genus "Stelis phaeoptera";
exd:local_conservation 0;
exd:blacklisted false .
exd:9 a exd:Species;
exd:genus "Arion vulgaris";
exd:common_name "Spanish slug";
exd:global_conservation 6;
exd:local_conservation 6;
exd:blacklisted true ;
exd:eats exd:10, exd:4 .
exd:10 a exd:Species;
exd:genus "Lupinus polyphyllus";
exd:common_name "garden lupin";
exd:global_conservation 6;
exd:local_conservation 6;
exd:blacklisted true .
exo:1 a exo:Observation; exo:species exd:1; exo:observed_time "2020-10-21T02:14:10"^^xsd:dateTime; exo:observed_lat 59.89; exo:observed_long 10.72 .
exo:2 a exo:Observation; exo:species exd:3; exo:observed_time "2021-01-01T02:14:10"^^xsd:dateTime; exo:observed_lat 59.39; exo:observed_long 10.78 .
exo:3 a exo:Observation; exo:species exd:5; exo:observed_time "2020-11-03T02:14:10"^^xsd:dateTime; exo:observed_lat 59.47; exo:observed_long 11.02 .
exo:4 a exo:Observation; exo:species exd:2; exo:observed_time "2020-01-17T02:14:10"^^xsd:dateTime; exo:observed_lat 58.91; exo:observed_long 10.34 .
exo:5 a exo:Observation; exo:species exd:6; exo:observed_time "2019-12-24T02:14:10"^^xsd:dateTime; exo:observed_lat 59.88; exo:observed_long 11.01 .
exo:6 a exo:Observation; exo:species exd:1; exo:observed_time "2020-06-09T02:14:10"^^xsd:dateTime; exo:location "Oslo" .
exo:7 a exo:Observation; exo:species exd:1; exo:observed_time "2021-01-02T02:14:10"^^xsd:dateTime; exo:observed_lat 59.81; exo:observed_long 9.94 .
exo:8 a exo:Observation; exo:species exd:10; exo:observed_time "2020-04-30T02:14:10"^^xsd:dateTime; exo:observed_lat 59.11; exo:observed_long 10.7 .
exo:9 a exo:Observation; exo:species exd:2; exo:observed_time "2019-02-12T02:14:10"^^xsd:dateTime; exo:observed_lat 59.36; exo:observed_long 11.08 .
exo:10 a exo:Observation; exo:species exd:1; exo:observed_time "2020-08-22T02:14:10"^^xsd:dateTime; exo:observed_lat 59.89; exo:observed_long 9.28 .
exo:11 a exo:Observation; exo:species exd:10; exo:observed_time "2020-10-20T02:14:10"^^xsd:dateTime; exo:observed_lat 60.01; exo:observed_long 10.78 .
exo:12 a exo:Observation; exo:species exd:9; exo:observed_time "2020-03-26T02:14:10"^^xsd:dateTime; exo:observed_lat 59.37; exo:observed_long 10.23 .
exo:13 a exo:Observation; exo:species exd:2; exo:observed_time "2020-09-04T02:14:10"^^xsd:dateTime; exo:observed_lat 59.41; exo:observed_long 10.4 .
exo:14 a exo:Observation; exo:species exd:1; exo:observed_time "2019-12-03T02:14:10"^^xsd:dateTime; exo:observed_lat 59.39; exo:observed_long 11.04 .
exo:15 a exo:Observation; exo:species exd:10; exo:observed_time "2020-10-06T02:14:10"^^xsd:dateTime; exo:observed_lat 59.11; exo:observed_long 10.83 .
exo:16 a exo:Observation; exo:species exd:2; exo:observed_time "2020-01-03T02:14:10"^^xsd:dateTime; exo:observed_lat 59.19; exo:observed_long 9.31 .
exo:17 a exo:Observation; exo:species exd:9; exo:observed_time "2020-07-07T02:14:10"^^xsd:dateTime; exo:observed_lat 59.64; exo:observed_long 10.61 .
exo:18 a exo:Observation; exo:species exd:10; exo:observed_time "2020-04-19T02:14:10"^^xsd:dateTime; exo:observed_lat 59.23; exo:observed_long 10.39 .
exo:19 a exo:Observation; exo:species exd:1; exo:observed_time "2019-03-08T02:14:10"^^xsd:dateTime; exo:location "Drammen" .
exo:20 a exo:Observation; exo:species exd:2; exo:observed_time "2021-01-21T02:14:10"^^xsd:dateTime; exo:location "Oslo" .
exo:21 a exo:Observation; exo:species exd:8; exo:observed_time "2020-11-30T02:14:10"^^xsd:dateTime; exo:observed_lat 58.99; exo:observed_long 9.78 .
exo:22 a exo:Observation; exo:species exd:9; exo:observed_time "2020-10-09T02:14:10"^^xsd:dateTime; exo:observed_lat 59.83; exo:observed_long 10.37 .
exo:23 a exo:Observation; exo:species exd:1; exo:observed_time "2020-07-23T02:14:10"^^xsd:dateTime; exo:location "Nord-marka" .exd:1 a exd:Species;
exd:genus "Rattus Norvegicus";
exd:common_name "Brown rat";
exd:global_conservation 6;
exd:local_conservation 6;
exd:blacklisted false .
exo:1 a exo:Observation;
exo:species exd:1;
exo:observed_time "2020-10-21T02:14:10"^^xsd:dateTime;
exo:observed_lat 59.89;
exo:observed_long 10.72 .can be represented as:
node:
hash | ntype | svalue
----------------------+-------------------------------------------+------------------------------------------------------
-3109702252271113716 | iri | http://example.org/species/obs/1
-4085280037482734459 | iri | http://www.w3.org/1999/02/22-rdf-syntax-ns#type
6099257321065622229 | iri | http://example.org/species/obs/Observation
7427117117437441385 | iri | http://example.org/species/obs/species
-5884675129065687946 | iri | http://example.org/species/desc/1
319924621413452125 | iri | http://example.org/species/obs/observed_time
1605665626047504580 | http://www.w3.org/2001/XMLSchema#dateTime | 2020-10-21T02:14:10
-8413395107724368320 | iri | http://example.org/species/obs/observed_lat
2596048708061934139 | http://www.w3.org/2001/XMLSchema#decimal | 59.89
1351448827599223571 | iri | http://example.org/species/obs/observed_long
-6079304262809190848 | http://www.w3.org/2001/XMLSchema#decimal | 10.72
2993837090040140756 | iri | http://example.org/species/desc/Species
5769607256422906705 | iri | http://example.org/species/desc/genus
-515250888949375450 | http://www.w3.org/2001/XMLSchema#string | Rattus Norvegicus
8858702909896475273 | iri | http://example.org/species/desc/common_name
6909716647617048169 | http://www.w3.org/2001/XMLSchema#string | Brown rat
-756248925033123473 | iri | http://example.org/species/desc/global_conservation
8015015005762445143 | http://www.w3.org/2001/XMLSchema#integer | 6
-5117885397842069576 | iri | http://example.org/species/desc/local_conservation
-3062248205756963312 | iri | http://example.org/species/desc/blacklisted
4351804349465540129 | http://www.w3.org/2001/XMLSchema#boolean | falsetriple:
subject | predicate | object
----------------------+----------------------+----------------------
-3109702252271113716 | -4085280037482734459 | 6099257321065622229
-3109702252271113716 | 7427117117437441385 | -5884675129065687946
-3109702252271113716 | 319924621413452125 | 1605665626047504580
-3109702252271113716 | -8413395107724368320 | 2596048708061934139
-3109702252271113716 | 1351448827599223571 | -6079304262809190848
-5884675129065687946 | -4085280037482734459 | 2993837090040140756
-5884675129065687946 | 5769607256422906705 | -515250888949375450
-5884675129065687946 | 8858702909896475273 | 6909716647617048169
-5884675129065687946 | -756248925033123473 | 8015015005762445143
-5884675129065687946 | -5117885397842069576 | 8015015005762445143
-5884675129065687946 | -3062248205756963312 | 4351804349465540129?s exd:blacklisted true;
exd:common_name ?name .The SQL-query becomes:
SELECT o2.svalue
FROM triples AS t1
JOIN nodes AS p1 ON (t1.predicate = p1.hash)
JOIN nodes AS o1 ON (t1.object = o1.hash)
JOIN triples AS t2 USING (subject)
JOIN nodes AS p2 ON (t2.predicate = p2.hash)
JOIN nodes AS o2 ON (t2.object = o2.hash)
WHERE p1.svalue = 'http://example.org/species/desc/blacklisted' AND
o1.svalue = 'true' AND
p2.svalue = 'http://example.org/species/desc/common_name';
?, e.g. ?x or
?personSELECT – Similar as in SQL (project and manipulate
values bound in query)WHERE – Match triples containing variables against
graphGROUP BY, HAVING, ORDER BY,
LIMIT, OFFSET – Similar as SQLPREFIX – introduces prefixes that can be used in
queryFROM not necessary, query evaluates over entire
graph!#)FILTER filters on valuesNULL-values in RDFOPTIONAL?x p/r ?y – same as ?x p [ r ?y ] .?x p|r ?y – matches ?x p ?y or
?x r ?y?x p+ ?y – ?x is related to
?y via one or more chained p properties
FROM-clauseWHERE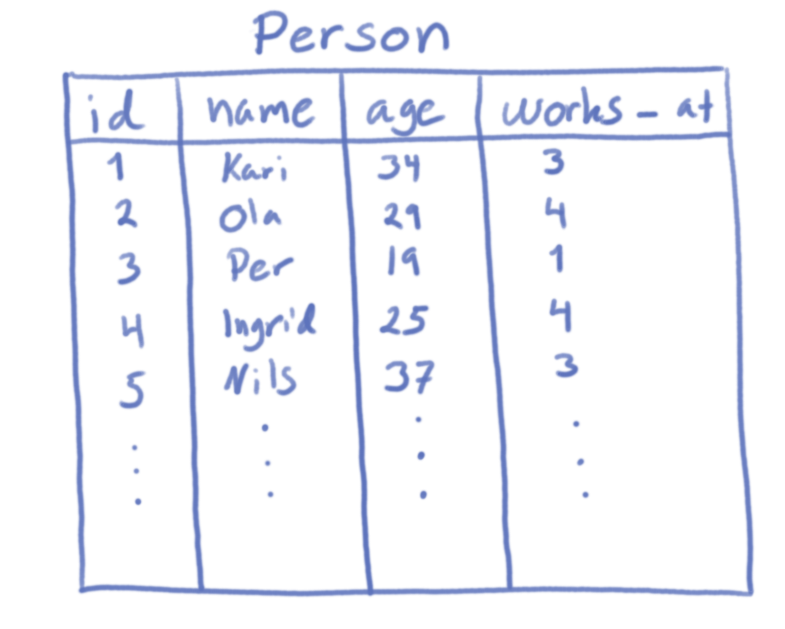



\d – lists tables/views etc.information_schema – tables describing everything in
the databaseDESCRIBE?s rdf:type rdfs:ClassSELECT-queries transform tables to tables
WITH,
etc.SELECT-queries transform graphs to tables
VIEW/WITH etc. in SPARQLCONSTRUCT-clause used in place
of SELECT
CONSTRUCT also contains a graph pattern (like
WHERE)CONSTRUCTResult:
exd:1 rdf:type exd:Species ;
rdfs:label "Rattus Norvegicus" .
exd:2 rdf:type exd:Species ;
rdfs:label "Larus Canus" .
exd:3 rdf:type exd:Species ;
rdfs:label "Lutra" .
exd:4 rdf:type exd:Species ;
rdfs:label "Osmoderna eremita" .
exd:5 rdf:type exd:Species ;
rdfs:label "Malacodea regelaria" .
exd:6 rdf:type exd:Species ;
rdfs:label "Balaenoptera physalus" .
exd:7 rdf:type exd:Species ;
rdfs:label "Emberiza calandra" .
exd:8 rdf:type exd:Species ;
rdfs:label "Stelis phaeoptera" .
exd:9 rdf:type exd:Species ;
rdfs:label "Arion vulgaris" .
exd:10 rdf:type exd:Species ;
rdfs:label "Lupinus polyphyllus" .PRIMARY KEYs)s p o g):
s p o gp o go g sg s pg po sINSERT/UPDATE/DELETECREATE/ALTER/DROPVIEWs for abstractionINSERT, DELETE, CREATE and
DROP also exists in SPARQL (no UPDATE!).ttl-file)GET, POST,
etc.) for manipulating triplestoresVIEWs!)In SPARQL we can pose queries impossible to write in SQL:
# What is the relationship(s) between Ole and Kari?
SELECT ?r
WHERE { ex:ole ?r ex:kari . }# Find all statements in DB
SELECT ?s ?p ?o
WHERE { ?s ?p ?o . }# Find all statements about Kari
SELECT ?p ?o
WHERE { ex:kari ?p ?o . }json (text) and jsonb
(binary)-> (get JSON-object) and ->> (get
field) to traverse the structure{"a" : 2, "b" : { "c" : 3 } } -> "b"
returns {"c" : 3}{"a" : 2, "b" : { "c" : 3 } } ->> "a" returns
2